

Protocol: The enzyme should not exceed 10 of total. NOT FOR USE IN DIAGNOSTIC PROCEDURES (EXCEPT AS SPECIFICALLY NOTED). Thermo Scientific NheI restriction enzyme recognizes GCTAGC sites and cuts best at 37☌ in Tango buffer (isoschizomers: AsuNHI, BmtI). Some enzymes may require additional DNA bases flanking the restriction site for complete digestion. In 2021, the market is growing at a steady rate and with the. Our mission is to develop high-quality innovative tools and services to accelerate discovery.įOR RESEARCH USE ONLY. The bacterial restriction-modification (R-M) system is made up of coordinated restriction and methyltransferase enzymes which are specific for a common DNA sequence. The Global Restriction Enzyme market is anticipated to rise at a considerable rate during the forecast period, between 20. As a member of the Takara Bio Group, Takara Bio USA is part of a company that holds a leadership position in the global market and is committed to improving the human condition through biotechnology. High Fidelity (HF) Restriction Enzymes have 100 activity in rCutSmart Buffer single-buffer.
provides kits, reagents, instruments, and services that help researchers explore questions about gene discovery, regulation, and function. NotI has a High Fidelity version NotI-HF (NEB R3189). NotI has a High Fidelity version NotI-HF ( NEB R3189 ). Tip: Methylation patterns also differ between different eukaryotes (see bullets above), affecting the choice of restriction enzyme for construction genomic DNA libraries.Takara Bio USA, Inc. Neb Reduce Star Activity with High-Fidelity Restriction Enzymes. Tip: Methylation patterns differ between bacteria and eukaryotes, so restriction patterns of cloned and uncloned DNA may differ. High-Fidelity (HF ®) restriction enzymes have the same specificity as native enzymes, but have been engineered for significantly reduced star activity and performance in a single buffer ( rCutSmart Buffer ).

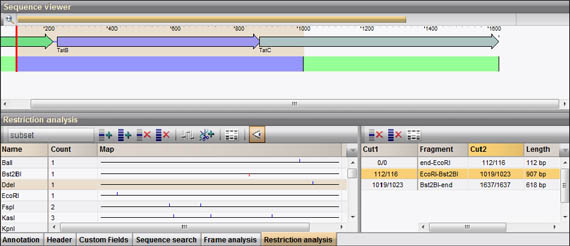
It is also used to quickly check the identity of a plasmid by diagnostic digest.
Plant DNA is highly methylated, so for successful mapping in plants, choose enzymes that either do not contain a CpG dinucleotide in their recognition site (e.g., DraI or SspI) or that can cleave methylated CpG dinucleotides (e.g., BamHI, KpnI, or TaqI). Restriction enzyme digestion is commonly used in molecular cloning techniques, such as PCR or restriction cloning. The simplicity of Anza restriction enzymes. Rare-cutter enzymes therefore cleave more frequently in these species. Drosophila, Caenorhabditis, and some other species do not possess methylated DNA, and have a higher proportion of CpG dinucleotides than mammalian species. Therefore many enzymes with CpG in their recognition site, such as EagI, NotI, and SalI, cleave mammalian DNA only rarely. The CpG dinucleotide occurs about 5 times less frequently in mammalian DNA than would be expected by chance, and most restriction enzymes with a CpG dinucleotide in their recognition site do not cleave if the cytosine is methylated. In addition, methylation patterns differ in different species, also affecting the choice of restriction enzyme. The type IIP restriction enzyme, NotI, from Nocardia otitidis-caviarum is a homodimer that recognizes the 8 bp DNA sequence 5-GC/GGCCGC-3 and cleaves both strands of DNA to create 5, 4 base cohesive overhangs ( Qiang and Schildkraut, 1987 ). Therefore the choice of restriction enzyme is affected by its sensitivity to methylation. The concentration of restriction enzymes is typically given as U/mL or U/L. In your reaction today, you should add 10 U of each enzyme to the reaction. It is suggested that you add a minimum of 5 U/g of DNA. During the plasmid DNA digest incubation, the NotI enzyme will specifically target and cleave the plasmid DNA. Restriction enzyme (s): In your table, you should have two different rows, one for each enzyme (NcoI-HF and NotI-HF). Not all restriction enzymes can cleave their recognition site when it is methylated. The Nuclease-free Water, 5X IVT Buffer, Supercoiled nonlinear plasmid DNA, and NotI restriction enzyme pre-conditioning entails pre-warming of all materials. Many organisms have enzymes called methylases that methylate DNA at specific sequences.







 0 kommentar(er)
0 kommentar(er)
